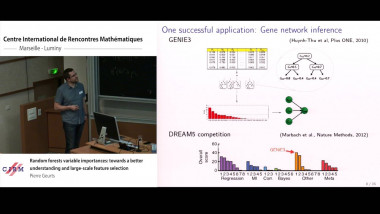
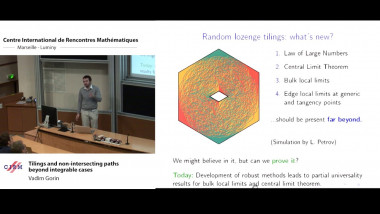
Random forests variable importances: towards a better understanding and large-scale feature selection
Also appears in collection : Thematic month on statistics - Week 1: Statistical learning / Mois thématique sur les statistiques - Semaine 1 : apprentissage
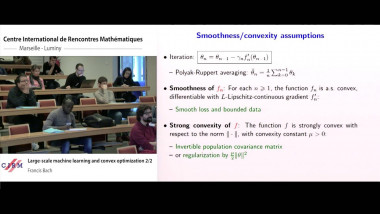
Random forests are among the most popular supervised machine learning methods. One of their most practically useful features is the possibility to derive from the ensemble of trees an importance score for each input variable that assesses its relevance for predicting the output. These importance scores have been successfully applied on many problems, notably in bioinformatics, but they are still not well understood from a theoretical point of view. In this talk, I will present our recent works towards a better understanding, and consequently a better exploitation, of these measures. In the first part of my talk, I will present a theoretical analysis of the mean decrease impurity importance in asymptotic ensemble and sample size conditions. Our main results include an explicit formulation of this measure in the case of ensemble of totally randomized trees and a discussion of the conditions under which this measure is consistent with respect to a common definition of variable relevance. The second part of the talk will be devoted to the analysis of finite tree ensembles in a constrained framework that assumes that each tree can be built only from a subset of variables of fixed size. This setting is motivated by very high dimensional problems, or embedded systems, where one can not assume that all variables can fit into memory. We first consider a simple method that grows each tree on a subset of variables randomly and uniformly selected among all variables. We analyse the consistency and convergence rate of this method for the identification of all relevant variables under various problem and algorithm settings. From this analysis, we then motivate and design a modified variable sampling mechanism that is shown to significantly improve convergence in several conditions.































































































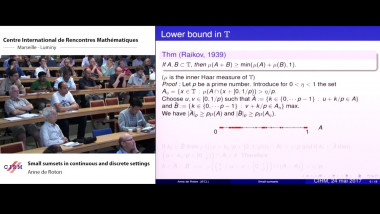
![$k$-sum free sets in $[0,1]$](/media/cache/video_light/uploads/video/2015-09-10_de_Roton-video--19cf70874d3c5af7378c268cab865b06.jpg)