Part 1 - Networks of prior knowledge as frames to understand complex biological data
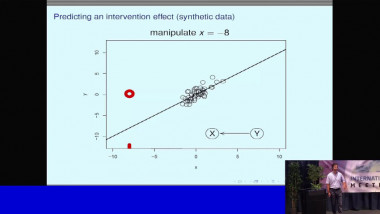
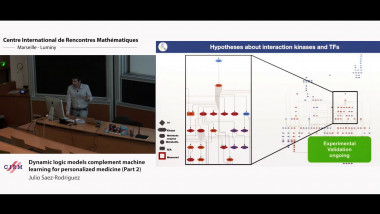
Modern technologies allow us to profile in high detail biomedical samples at fast decreasing costs. New technologies are opening new data modalities, in particular to measure at the single cell level. Prior knowledge, and biological networks in particular, are useful to integrate this data and distill mechanistic insight. This can help to interpret the result of machine learning or statistical analysis, as well as generate input features for these methods. In addition, they can be converted in dynamic mechanistic models to gain more specific insight. I will give an overview of these approaches showcasing some examples and approaches used in the field.













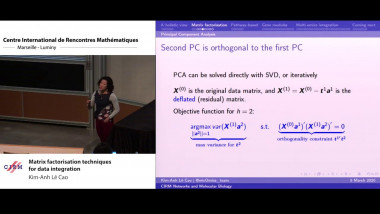
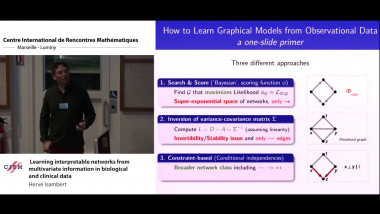
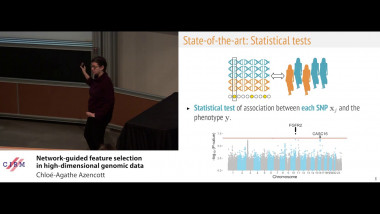
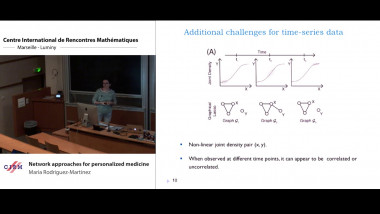

![[1243] Degrés dynamiques](/media/cache/video_light/uploads/video/SeminaireBourbaki.jpg)