Part 2 - Dynamic logic models complement machine learning for personalized medicine
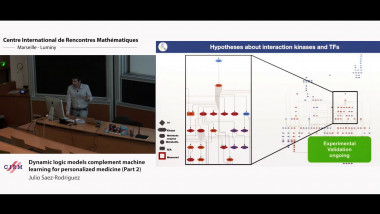
In the second talk, I will present some of our work on this area. Our work on this area, where we have focused on transcriptomics and (phospho)proteomics to study signaling networks. Our tools range from a meta-resource of biological knowledge (Omnipath) to methods to infer pathway and transcription factor activities (PROGENy and DoRothEA, respectively) from gene expression and subsequently infer causal paths among them (CARNIVAL), to tools to infer logic models from phosphoproteomic and phenotypic data (CellNOpt and PHONEMeS). We have recently adapted these tools to single-cell data. I will illustrate their utility in cases of biomedical relevance, in particular to improve our understanding of cancer and to develop novel therapeutic opportunities. As main application I will discuss our work analysing, as a model for personalized medicine, large pharmaco-genomic screenings in cell lines. These screenings provide rich information about alterations in tumours that confer drug sensitivity or resistance. Integration of this data with prior knowledge provides biomarkers and offer hypotheses for novel combination therapies. Our own analysis as well as the results of a crowdsourcing effort (as part of a DREAM challenge) reveals that prediction of drug efficacy from basal omics data is that discussed above is far from accurate, implying important limitations for personalised medicine. An important aspect that deserves detailed attention is the dynamics of signaling networks and how they response to perturbations such as drug treatment. I will present how cell-specific logic models, trained with measurements upon perturbations, can provides new biomarkers and treatment opportunities not noticeable by static molecular characterisation.














![[1243] Degrés dynamiques](/media/cache/video_light/uploads/video/SeminaireBourbaki.jpg)