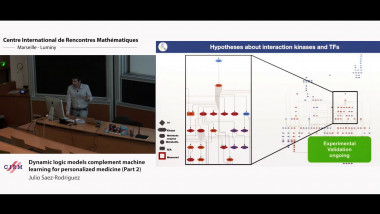
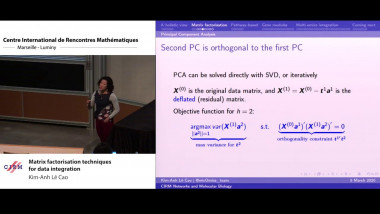
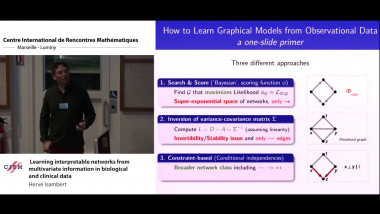
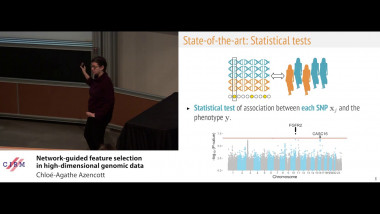
Matrix factorisation techniques for data integration
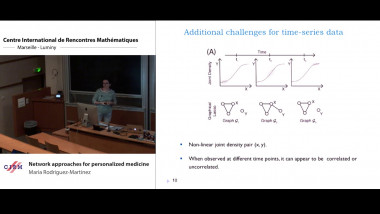
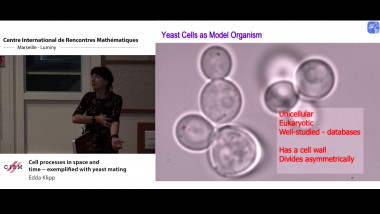
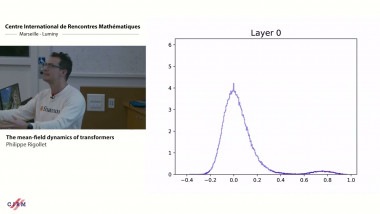
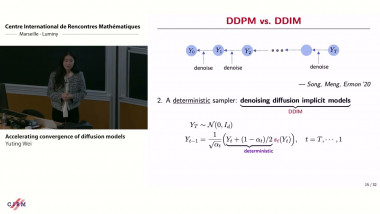
Gene module detection methods aim to group genes with similar expression profiles to shed light into functional relationships and co-regulation, and infer gene regulatory networks. Methods proposed so far use clustering to group genes based on global similarity in their expression profiles (co-expression), bi-clustering to group genes and samples simultaneously, network inference to model regulatory relationships between genes. In this talk I will focus on multivariate matrix decomposition techniques that enable dimension reduction and the identification of molecular signatures. We will consider two different types of assays: bulk and single cell assays. Bulk transcriptomics assays use RNA-sequencing techniques to monitor the average expression profile of all the constituent cells, but fail to identify the distinct transcriptional profiles from different cell types. Single cell assays use similar RNA-seq techniques (scRNA-seq) to those used for bulk cell populations, but provide unprecedented resolution at the cell level to understand cellular heterogeneity and uncover new biology. However, scRNA-seq present new computational and analytical challenges, because of their sheer size (100K – 500K of cells are sequenced) and their zero inflated distribution due to technical drop-outs. I will illustrate how we can use matrix factorisation technique to mine these data and identify gene modules that underpin molecular mechanisms in cell identity in scRNA-seq. I will also give further perspective on how we could extend similar concepts to integrate different omics data types (e.g. bulk transcriptomics, proteomics, metabolomics) to identify tightly connected multi-omics signatures that holistically describe a biological system.