Evolutionary ecology of antibiotic resistance
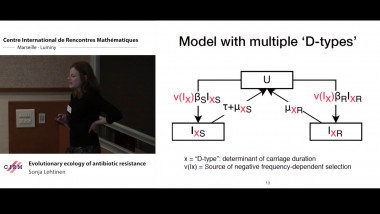
Antibiotic resistance is a serious public health concern. Responding to this problem effectively requires characterising the factors (i.e. evolutionary and ecological processes) that determine resistance frequencies. At present, we do not have ecologically plausible models of resistance that are able to replicate observed trends - we are therefore unable to make credible predictions about resistance dynamics. In this talk, I will present work motivated by three tends observed in Streptococcus pneumoniae resistance data: the stable coexistence of antibiotic sensitivity and resistance, variation between resistance frequencies between pneumococcal lineages and correlation in resistance to different antibiotics. I will propose that variation in the fitness benefit gained from resistance arising from variation in the duration of carriage of pneumococcal lineages is a parsimonious explanation for all three trends. This eco-evolutionary framework could allow more accurate prediction of future resistance levels and play a role in informing strategies to prevent the spread of resistance.