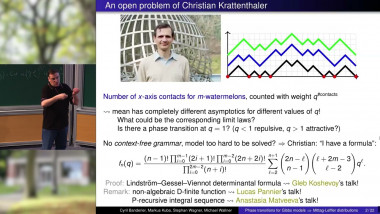
Appears in collection : Stochastic Processes in Evolutionary Biology / Processus Stochastiques en Biologie Evolutive
Consider two ancestral lineages sampled from a system of two-dimensional branching random walks with logistic regulation in the stationary regime. We study the asymptotics of their coalescence time for large initial separation and find that it agrees with well known results for a suitably scaled two-dimensional stepping stone model and also with Malécot's continuous-space approximation for the probability of identity by descent as a function of sampling distance.
This can be viewed as a justification for the replacement of locally fluctuating population sizes by fixed effective sizes. Our main tool is a joint regeneration construction for the spatial embeddings of the two ancestral lineages.