General epidemiological models: law of large numbers and contact tracing
Apparaît également dans la collection : 5th Workshop Probability and Evolution / 5ème rencontre Probabilités et évolution
We study a class of individual-based, fixed-population size epidemic models under general assumptions, e.g., heterogeneous contact rates encapsulating changes in behavior and/or enforcement of control measures. We show that the large-population dynamics are deterministic and relate to the Kermack-McKendrick PDE. Our assumptions are minimalistic in the sense that the only important requirement is that the basic reproduction number of the epidemic $R_0$ be finite, and allow us to tackle both Markovian and non-Markovian dynamics. The novelty of our approach is to study the "infection graph" of the population. We show local convergence of this random graph to a Poisson (Galton-Watson) marked tree, recovering Markovian backward-in-time dynamics in the limit as we trace back the transmission chain leading to a focal infection. This effectively models the process of contact tracing in a large population. It is expressed in terms of the Doob h-transform of a certain renewal process encoding the time of infection along the chain. Our results provide a mathematical formulation relating a fundamental epidemiological quantity, the generation time distribution, to the successive time of infections along this transmission chain.












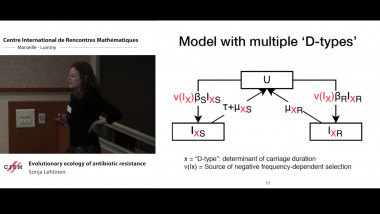
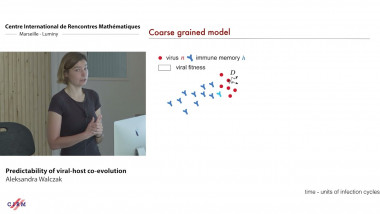



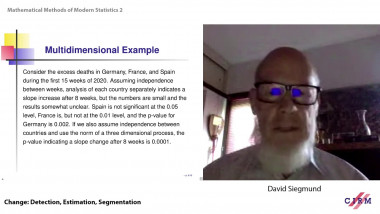







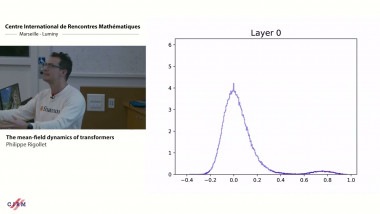
![[1243] Degrés dynamiques](/media/cache/video_light/uploads/video/SeminaireBourbaki.jpg)
